Sciencepic of the day (3): Protein Detection by means of Western Blotting
Todays' science pic comes from an experiment in my lab. It has a flaw (more about that later), but it serves well to make some points and explain the technology. Stay with me; this post is a bit longer as I thought it would be.
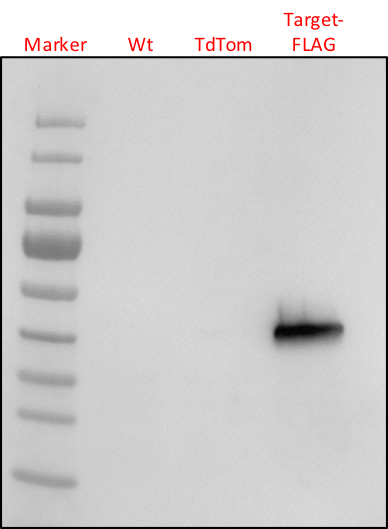
SciencePic: Western Blot based detection of a Flag-tagged protein in Phytophthora capsici lysates. Source: My own collection.
Molecular biologists love to study the function of proteins in organisms. One important way by doing so is to either change the levels of a given protein in cells or whole organisms (in vivo studies), express and purify the protein for studies in test tubes (in vitro) or determine structures. Irrespective of the approach or purpose, a researcher always has to ask her/himself the question: is my protein present and if so at what levels? One way of doing so is to probe for protein levels, and one can do so by doing a Western Blot. The principle of a Western is simple. First, proteins need to be separated. This is done by size and overall charge by migrating a sample through a polyacrylamide gel that has tiny pores the protein can travel through. It is called gel electrophoresis Along with your set of samples (one sample is typically loaded onto one lane), we run a ladder (a commercially produced sample containing a mix of proteins with known sizes) to estimate the size of your protein. We call them markers. After gel separation; we transfer the proteins to a membrane (We refer to this as blotting), and the proteins present on the membrane we then probe with antibodies that are either specific to the protein of interest (these are often custom made) or target a known sequence motif or domain. These are called epitope, and the proteins, therefore, needs to be epitope-tagged. In the case of the science pic, we used a FLAG-TAG.
After probing, we need to detect the protein. We do this using antibodies (often called secondary antibodies) that carry an enzyme (for example, horseradish peroxidase). These enzymes convert a substrate we throw on the blot and produces a visible signal (light). This light signal can be captured, measured and graphically represented in an image (as shown here).
Now, this image shows an experiment in which we try to detect a protein of interest in P. capsici. Four samples have been loaded (from left to right). Our size marker, two negative controls (a wildtype strain and a transgenic strain expressing a protein that does not have a FLAG-TAG) and our strain of interest that may or may not express our Target-FLAG protein.
As you can see, there is a clear band that is specific to our strain of interest. Nothing shows up in the negative control samples, indicating that the antibody is detecting our protein of interest. Because of the marker, we can also asses the approximate size of our protein of interest. In this case, it matches our expected size. We, therefore, have good evidence that our protein of interest is expressed. There is, however, a caveat. One conclusion could be that the band is specific and reflects the expression of our precious target protein. It's the results we want. An alternative explanation (hypothesis) is that the signal is aspecific, but that this signal is absent because of compromised negative control samples. What if the negative controls did not have any proteins? Or lower levels? This would also explain the absence of a band and make our result look specific. One way to eliminate this possibility is to use loading control. This involves loading the same samples on a separate gel and staining all proteins afterward or by staining the blot that we probed with our antibody. This control was not performed in this experiment, unfortunately. We, therefore, cannot use it for publication.
Links of interest:
https://en.wikipedia.org/wiki/Western_blot
https://en.wikipedia.org/wiki/Gel_electrophoresis_of_proteins
https://en.wikipedia.org/wiki/Antibody
I hope you will find these pieces interesting. Your vote and follow help me open up the exciting world of scientific research to you.
Thank you @foundation for this fantastic SteemSTEM gif

You flag a protein!!! That must be a spam protein hahahahahahaha
let me see if i can get an upvote tag protein XDDD
Haha in some ways it is. If you are willing to consider pathogens as spammers!
Its very nice and good post i like it i upvoted
For you plz upvote me
https://steemit.com/photography/@keerthi12345/photography
"Protein Control" -Prince
Further probing needed.
Hi. Thank you for your comments. Not sure what exactly you are trying to suggest to me though. Happy to elaborate if you could. 👍🏼