Lab Diaries #2 - Making My Own Transgenic Cell Line
In the previous post of this series I explained the significance of using cell lines in cancer research. Today I would like to continue by sharing some details about my research, and to show you how interesting and challenging at the same time science actually is!
Missing an adequate model system for your research? Make one!
The idea of our project was to investigate transcriptional and translational changes in Multiple Myeloma cell line induced by over-expression of particular oncogene.
However, to be able to properly assess changes going on in your cells, you need to have cell line that has high levels of gene, and the SAME cell line with low levels of that gene, so you could compare them and eventually attribute all observed changes to high expression of your gene of interest, because everything else must stay the same.
But, where to find cell line that has both low and high expression of the same gene? It doesn't exist, of course!
This means that you have to make your own model system, a cell line that will have high or low levels of whichever gene you need.
So I did it!!! Want to know how? ;)
Making my own transgenic cell line by lentiviral transduction
For our experiments we decided to use a cancer cell line which has by default low expression of our gene on interest, and to create a transgenic cell line, which will be the same as parental line, except for having high expression of our gene of interest.
To do that, you need to insert your gene somehow into parental cell line, and to force that cell line to produce a protein product of that gene, in other words - you need to have a stable expression of your gene of interest to be able to do experiments with such cell line.
This is achieved by using lentiviral vectors in a process called lentiviral transduction.
Lentiviruses are subtype of retroviruses (viruses with RNA genomes) and are specific by the fact that they can infect both non-dividing and actively dividing cells, unlike retroviruses, which can infect only mitotically active cells. HIV virus is an example of lentivirus.
Lentiviral vectors are basically lentiviruses that are modified in such way that your gene of interest is inserted into their genome, and they are capable of infecting your cell line, deliver your gene which becomes stably integrated into the genome of your cell line, BUT they do not have the capability of reproducing themselves, in other words, they cannot form new viral particles and infect other cells.
Once lentiviral vector integrates your gene into the genome of your cell line, that gene is being transcribed into mRNA and translated into protein product, in other words - you got yourself a transgenic cell line that produces your protein of interest like crazy!
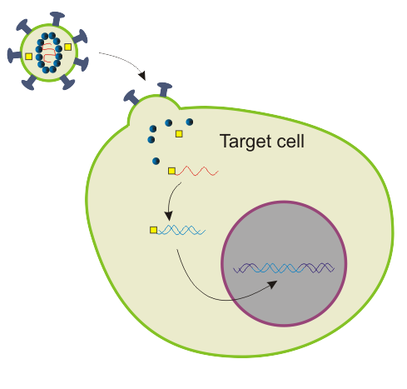
Modified from Wikimedia Commons, public domain
One batch of cells was transfected with lentiviruses carrying control vectors - vectors without our gene of interest (GOI), and the other batch was transfected with lentiviruses that contained our GOI. Vectors also contained virus-specific genes necessary for integration of vector into genome of human cells, and very important gene for antibiotic (in this case puromycin) resistance.
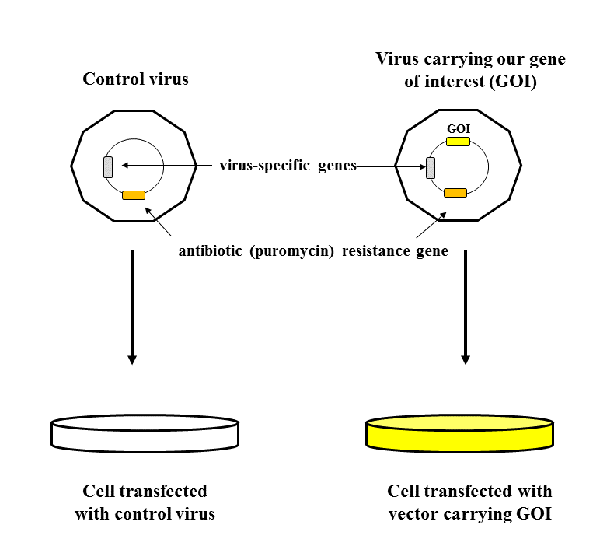
Why do we need gene for antibiotic resistance? Because it represents selection marker, based on which we are able to tell which cells were successfully transfected, and which did not integrate the vectors.
Selection is made simply by growing cells in selective media, which contains antibiotic (puromycin). Cells that were successfully transfected will continue to grow in media containing puromycin, whilst cells that did not take up the plasmid will die, because they don't have puromycin resistance and the antibiotic will kill them.
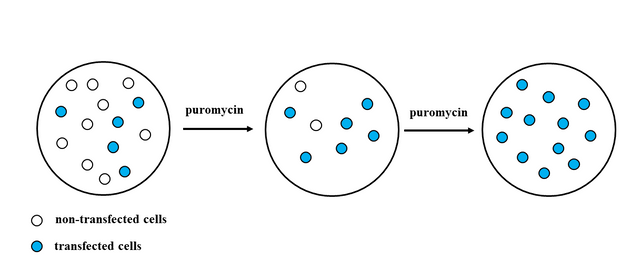
After you have confirmed that you have uniform population of cells (cells that have taken up the plasmid), you have to perform qPCR and Western Blot experiments with your transgenic cell line to confirm that you have stable expression of your GOI .
When you see amplification in qPCR and get bands in Western Blot - congratulations, you have created your model system successfully!!!
You can finally proceed to your experiments using your cells... and this is where you realize that the party has just begun :)
In the next Lab Diaries post I will write about the experiments and methods I use in my research.
Until then, relax and keep steemSTEM! ;)
Literature
Protocol for Lentiviral Transduction of Human Cells
Image credits pixabay.com and myself, unless otherwise stated
For more scientific-related content check steemSTEM. Follow me if you like my posts and want to read some more ;) If you have any thoughts/suggestions fell free to leave a comment!
Being A SteemStem Member
Nice write up@scienceangel! When the lentivirus integrates your gene of interest into the genome is it random or a targeted locus? If it is random could there be non specific consequences of where it integrated and not necessary from the over-expression of your gene of interest? Can I ask which oncogene you over-expressed?
Cheers!
Love Genetics!
ian
Interesting seeing you here :D
I love science and I am sure you do to!
Absolutely ^^ I hope you follow scienceangel, she's great! I can also highly recommend @irime , she doesn't post much but all her posts are top quality!
Yes I follow @scienceangel and thanks for the recommendation to follow @irime. I also follow @alexander.alexis as he has some very comprehensive posts that are meant for the general public.
Thank you, and thanks for very interesting questions! We do not know exactly where our gene of interest is integrated, but we know that the process of integration site selection is not random, and that lentiviruses most often integrate into actively transcribed genes, because other parts of genome are "wrapped up'' within heterochromatin. In theory, there could be some "undesired" consequences like virus integrating within the exon of a gene, disrupting its activity, but in such cases there's always another copy of the gene left. The project I'm writing about is still ongoing, so I cannot reveal details until we publish our results :)
I guess you can get multiple independent lines to correct for non specific effects. Have you considered using CRISPR Cas9 to target your gene to a specific locus? I totally understand about not mentioning the name of your over-expressed gene. Good luck with your research!
CRISPR/Cas9 is used for targeted genome editing in such way that you perform knockout of the gene of interest. It cannot be used to "direct" lentiviral integration to the specific site (at least I haven't heard of that).
Here is a nice review on some of the CRISPR applications and tools
Thanks for the review, but I wasn't able to find application of CRISPR/Cas9 where you would make the cut and integrate your gene at specific site in genome by using only CRISPR/Cas9. If you would be so kind to point out on the exact paragraph which describes such application, because I would really like to learn something new! :)
Yes its called Homology Directed Repair (HDR) and its in Figure 1B of that review but here is a figure from NEB
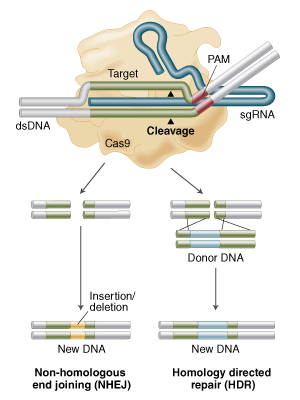
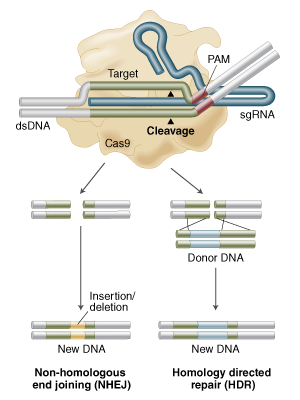
showing Non Homologous End Joining (NHEJ) for knock outs and HDR for "knocking in" any piece of DNA you can dream of (the donor DNA with homology arms in figure below).
Also have a look at this video as you might be interested in using HDR in non dividing cells:
Great, I actually didn't know about this particular application of CRISPR! However, after searching through PubMed for publications/applications, I saw that method is still in its early phase of development, and that it still doesn't exhibit great efficiency. But thanks anyway for sharing this, I will certainly follow further development of this method!
I am almost certain that whatever your end result you wanted using your lentiviral integration you can do with CRISPR Cas9 technology, but the beauty of this is that the integration is not random. CRISPR is not just for knock outs of genes. For example in your research if you want a strong promoter driving your gene of interested (like your lentivirus vector) you just make the construct and direct CRISPR Cas9 where to make the cut and integration site of your gene of interest.
I had to make a model system for my honors project, years ago! Unfortunately of my alloted 12 months for the project, the yeast wouldn't take their damn genes for 6 of them :/
So sorry to hear that! How did you make yeast cells to take the genes at the end? For human cancer cells, I added polybrene into the growth media, it should increase the efficiency of transduction... I actually managed to make my transgenic cells after the first try, maybe it was beginner's luck :)
In the end, my labmate got it down, I can't remember the specifics it was so long ago. We had an endless array of reasons it never worked... mistakes, contaminations, mistakes, bad lucks... mistakes.
It's awesome when things work the first time around! Well done :)
ha ha imagine if you working with mice instead of yeast. It seems easy on paper put in practice there are so many things that can go wrong.
Yeah, I can imagine that'd be a nightmare!
Lovely discovery, i always thought it was impossible or difficult to be in possession of one individuals transgenic cell line, i will have to share this with my friend from the field who argues alot. Thanks for sharing
Hello, as a member of @steemdunk you have received a free courtesy boost! Steemdunk is an automated curation platform that is easy to use and built for the community. Join us at https://steemdunk.xyz
Upvote this comment to support the bot and increase your future rewards!