Setting up R Environment for the Command Line
I am pretty sure that I am one of the few R users that prefer to use R at the command line over the very popular RStudio interface. Though there are many holy wars over silly developer stuff, this is not one that I'm willing to fight or die on a hill for nor is this post a persuasive piece on why you should use command line R over a GUI package.
However, for those who are using R through command line/terminal either out of preference or necessity, here are some customization options I’ve learned that may help speed things along.
Finding R_HOME
The directory where R environment and profile settings are stored can be found through using R.home() inside the R console. This is useful because many of the files governing the R start is in the ‘R_HOME/etc/’ folder.
There are two files that we are mainly concerned with: Renviron.site and Rprofile.site. Renviron contains general environmental variables such as where the installation path to your R libraries are kept and allows for the definition of other environment variables that might be used in deeper levels of integration between R and the operating environment.
Rprofile
Rprofile allows for the customization of option sites, package loading, and other quality of life settings that makes R a lot more comfortable to use in terminal. For example, two of the options I used to always run at the top of my code is as follows:
options(scipen=20)
options(stringsAsFactors = FALSE)
By default, R imports character strings in data.frames as factors. This becomes exceptionally annoying when your data set comes in multiple files, and combining these data sets into one large data frame requires you to manually convert all factors into strings. My personal preference is to always keep strings as characters unless I really am either in need of the memory savings or am definitively sure that my array contains the entire universe of permitted values for that data element.
By appending these options to the end of Rprofile, I save myself the silliness of either retyping or source my option sets each time I start a new project.
Word of caution: Environmental options sometimes make code more difficult to replicate, as someone that is running your script may not have the same settings. Always publish relevant settings when sharing code with others!
Add Some Color (Linux)
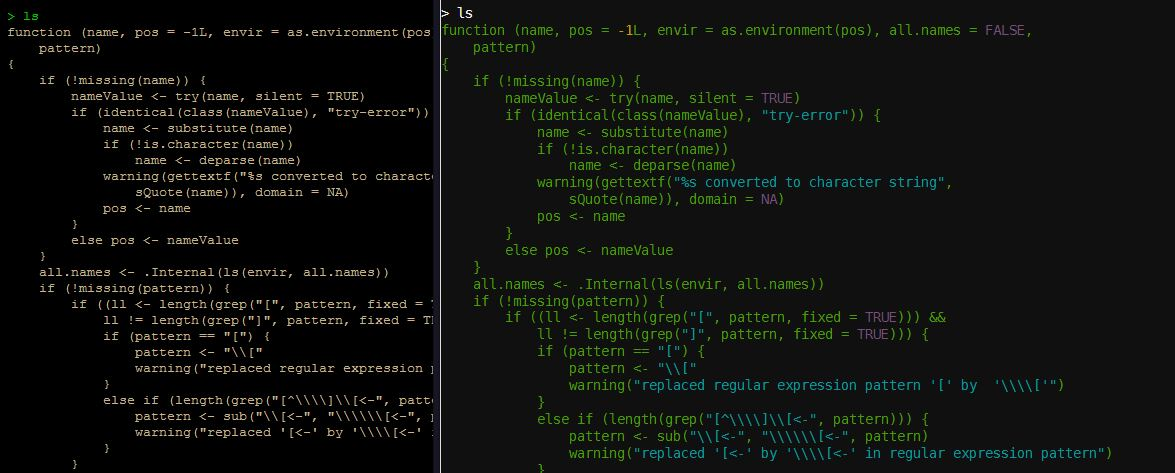
Alas, this next option is only for the Linux/Unix flavor of R but the colorout package adds syntax highlighting and a bunch of other goodies to the plain grey R console that we’ve come to love and squint at. Below, I show you a before and after version of using colorout in looking at the function ls():
Awesome right? 'colorout' can easily be installed from github using devtools by typing: devtools::install_github("jalvesaq/colorout")
As icing to the cake, colorout, or any package for that matter, can be loaded by default by adding the following code to Rprofile:
options(defaultPackages=c(getOption("defaultPackages"), "colorout", "other_packages..."))
Thanks for the tutorial. Even if python^tm is better. /s